Kernel provides a set of analysis tools to give you a quick glance at your datasets to verify that tasks were presented correctly, and otherwise aid in your research. These tools are accessed in the Pipelines tab for each dataset. For instructions on downloading an analysis file, see Downloading datasets.
Kernel Analysis Tools
Analysis: Task
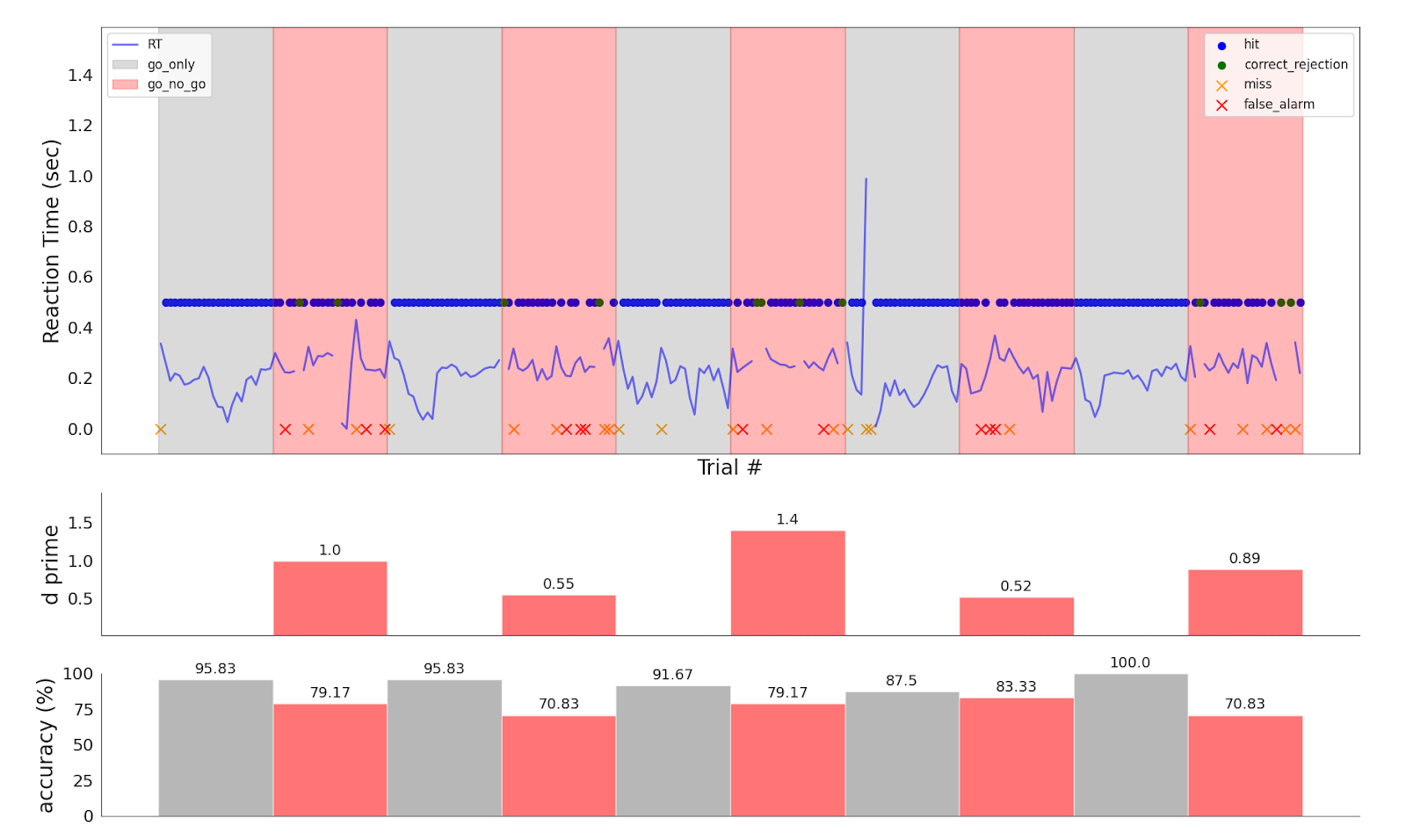
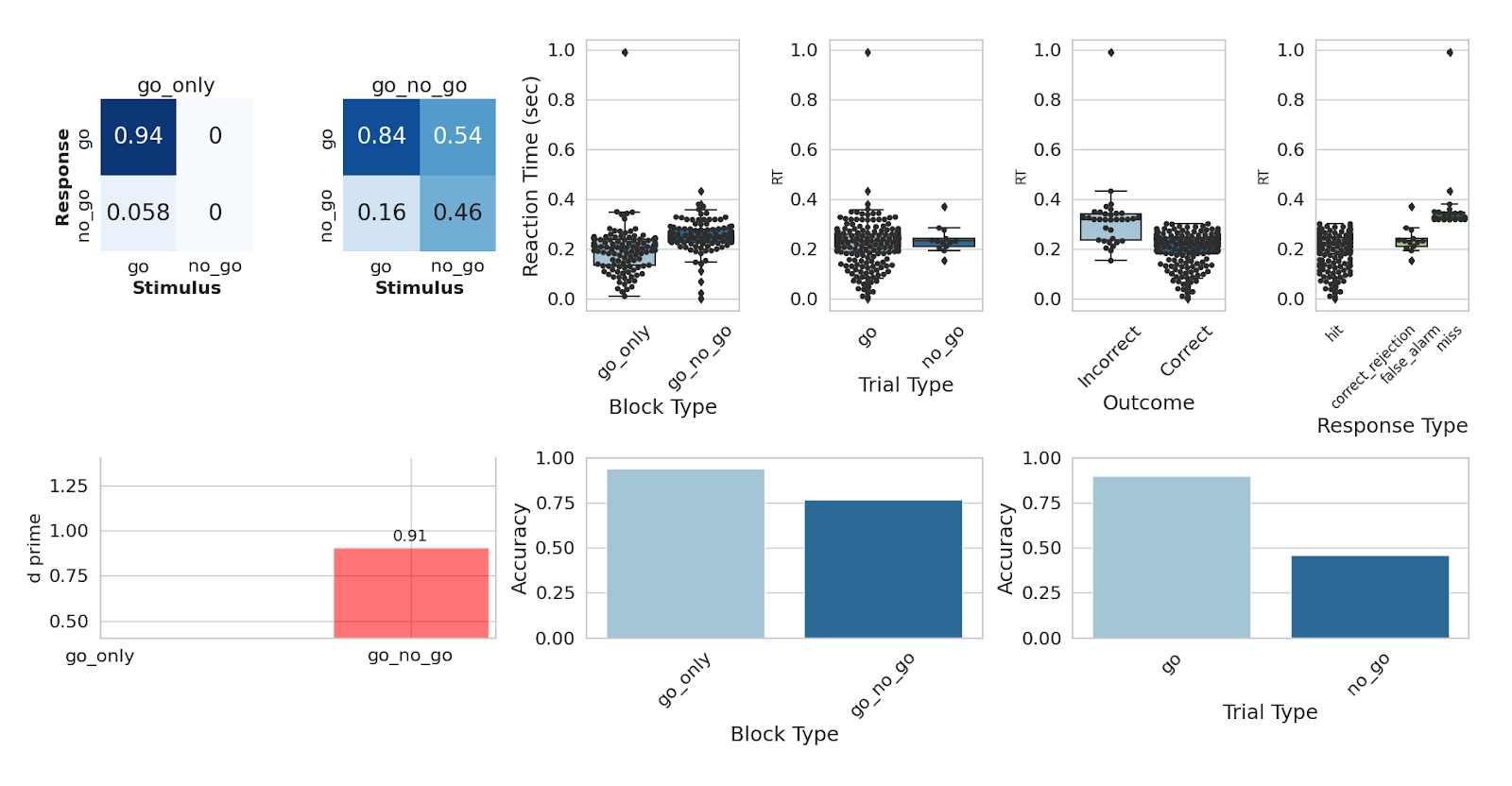
These figures are an example output from a behavioral analysis run on Go/No-Go data. You can expect to see summary metrics of the task-specific behavioral data, including reaction time, accuracy, and response classification; and also distribution of the timing of different events in the task, to check that the task was presented correctly.
NIRS: General Linear Model
The General Linear Model (GLM) is a statistical technique used to analyze hemodynamic signals, originally developed for functional Magnetic Resonance Imaging (fMRI) and subsequently adopted by the functional Near-Infrared Spectroscopy (fNIRS) community. Kernel applies preprocessing to the data before GLM statistical analysis, identical to the preprocessing in the SNIRF: Hb Moments pipeline.
Design Matrix
The design matrix for the GLM, shown in the Analysis report, typically includes:
- Regressors for Main Conditions: These are experiment-specific and represent the main conditions in the experiment.
- Trend-Capturing Regressors: A set of discrete cosine transform basis functions used to capture trends in the data.
- Constant Term: A baseline constant term.
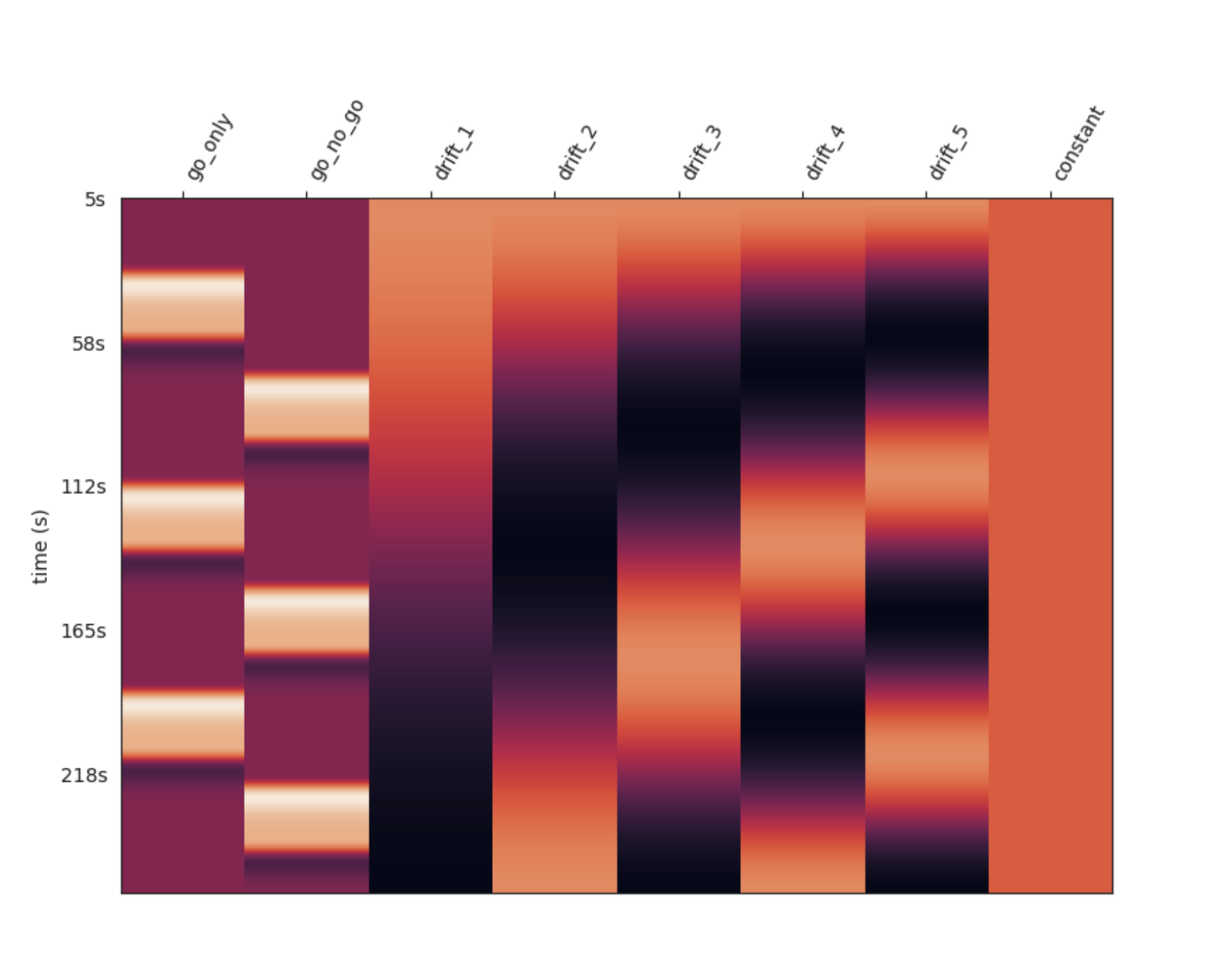 Example Design Matrix
Example Design MatrixInterpretation
This plot is necessary to interpret the following statistical results — it indicates how the data was modeled and how conditions were defined for statistical inference.
Contrasts
Contrasts (typically T-contrasts) are formed between regressors of interest. An interactive topoplot is created to visualize the effect sizes, p-values, and T-statistics for each channel.
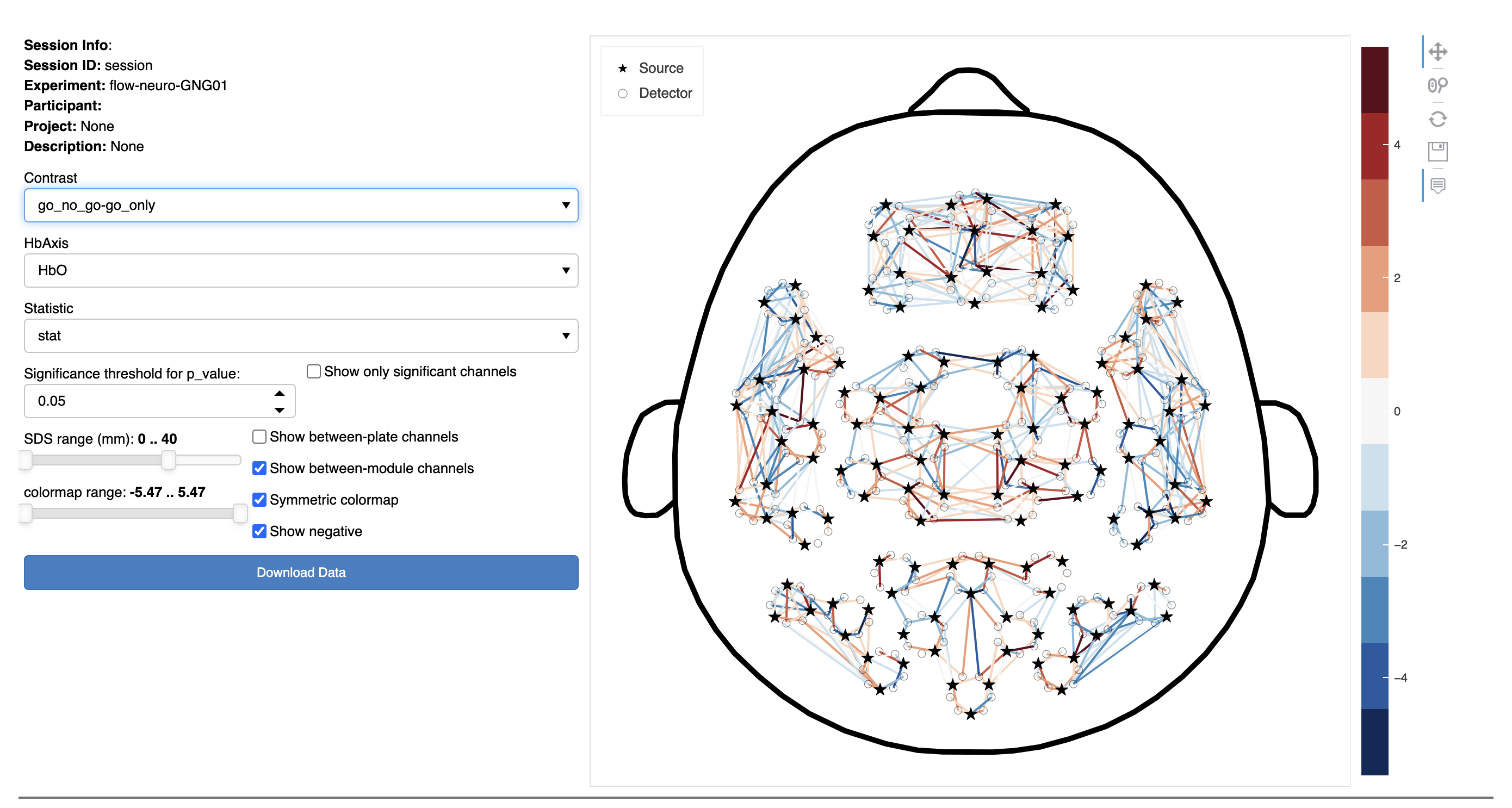
In this interactive figure, the statistical results of the GLM analysis are shown using a top view of the head and a schematic channel layout.
The plot on the right represents source-detector (SD) pairs as colored lines between the corresponding source (stars) and detector (circles). The shading of these lines are defined according to the colormap range. Hover over a line to show the channel name and associated value.
On the left side of the figure, there are controls to dynamically customize the data visualized:
- Contrast: Select the experimental contrast of interest (i.e., statistical comparison between conditions). For a given experiment, several contrasts may be available.
- Hb Axis: Choose a chromophore (HbO or HbR)
- Statistic: Chose from:
- Theta weight (effect size)
- Stat (this may be a F-statistic or a t-statistic for the GLM contrasts)
- p-value
- Significance threshold for p_value: Select a p-value value level above which channels will be ignored.
NOTE: this setting is ignored unless the Show only significant channels checkbox is selected. - Show only significant channels: Select this checkbox to limit the display to only statistically significant channels based on the threshold specified.
- SDS Range: Only show channels above (the left control) and below (the right control) the number selected.
- Colormap Range: Set the lower and upper limits of the color map (to optimize the display).
- Show between-plate channels: Select to include channels where the source and detector are on different headset plates.
- Show between-module channels: Select to include channels where the source and detector are on different modules.
- Symmetric colormap: Makes the color representation symmetrical. It is usually desirable for the maximum negative and positive values of the scale to match. Uncheck this option to remove that constraint.
NOTE: Selecting this affects the Colormap Range slider (the maximum bound of the color map is mirrored). - Show negative: Select to include negative values based on the option selected in the Statistic pop-up menu above.
To download the results from the analysis in a .csv format:
- Click the Download Data button at the bottom left of the figure.
NIRS: Epoched
Epoched analysis offers a straightforward method for examining results by segmenting the data around events of interest and analyzing the average signal by condition. This approach is suitable for experimental designs featuring long rest periods between blocks or trials, allowing the hemodynamic response to return to baseline before the next condition. For fast event-related designs, the GLM is required.
Before epoching the data, additional preprocessing steps are performed. The steps below follow the processing steps in the SNIRF: Hb Moments pipeline.
9. High Pass Filtering
This step involves moving average detrending with a sliding window of 100 seconds.
10. Low Pass Filtering
A Finite Impulse Response (FIR) filter is used, with a cutoff frequency of 0.2 Hz, a width of 0.1 Hz, and a ripple of 40 (not too steep to avoid ringing artifacts and phase distortion). This filters out heart rate related fluctuations and high-frequency noise to improves statistical outcome.
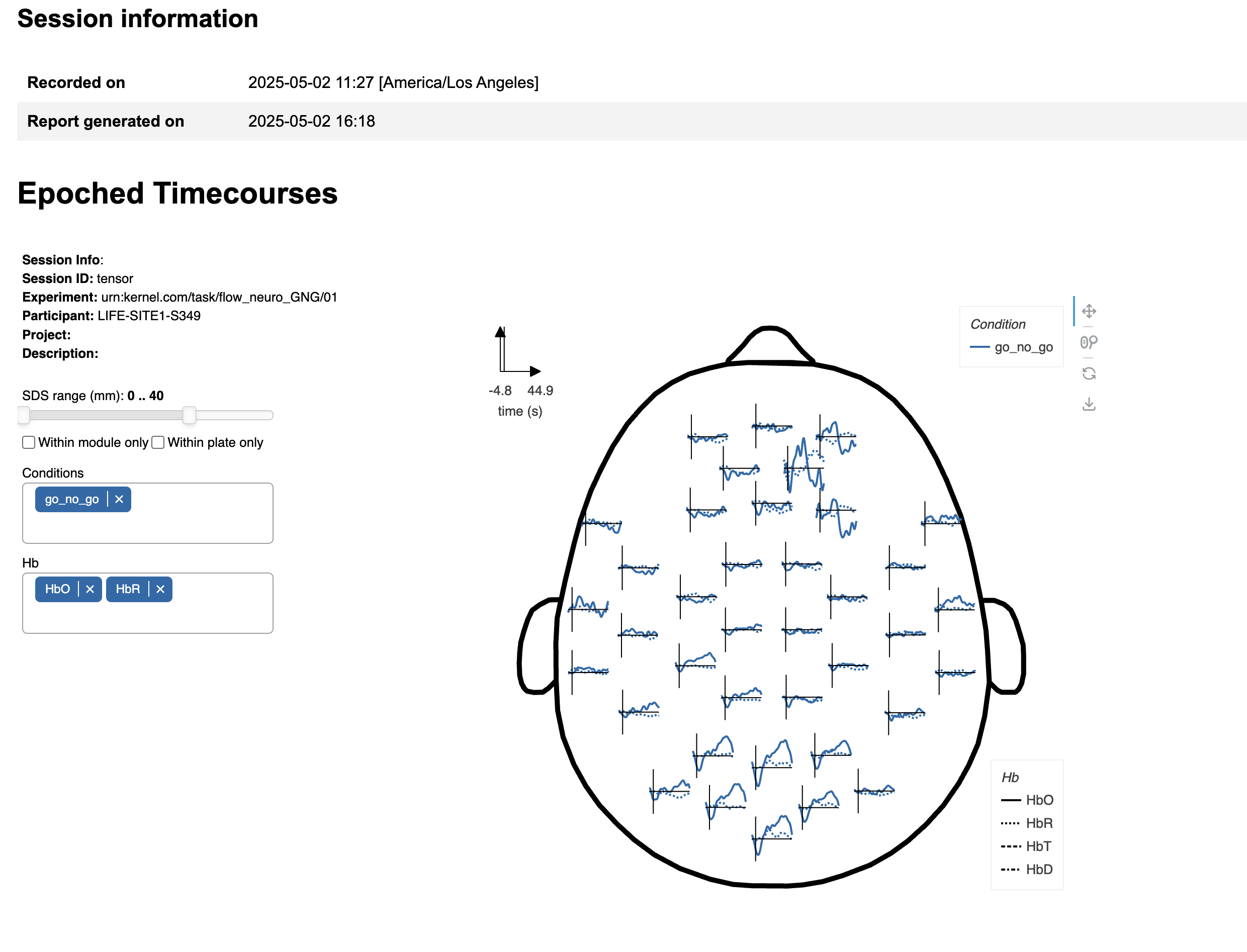
This interactive figure displays the results of the epoched analysis from a top view of the head, using a schematic layout of the modules. Each line graph represents the average timecourse of the selected hemoglobin signal (e.g., HbO, HbR) across all channels terminating on the module corresponding to the plot's location.
On the left side of the figure, there are controls to dynamically customize the data visualized:
- SDS Range: This slider filters channels by source-detector separation (SDS). By default, all channels with SDS ≤ 40 mm are included, spanning within-module, inter-module, and cross-plate connections.
- Within module only: Restricts analysis to channels where both the source and detector are located on the same module ( SDS ≤ 26 mm).
- Within plate only: Restricts channels to those within and between modules only on the same physical plate. Inter-plate channels typically have longer SDS. See a map of the plates here.
Conditions: Select one or more experimental conditions to visualize. Available conditions depend on the specific experiment run.
Hb Axis: Choose which hemoglobin signal(s) to display:
- HbO: Oxygenated hemoglobin
- HbR: Deoxygenated hemoglobin
- HbT: Total hemoglobin (HbO + HbR)
- HbD: Hemodynamic difference (HbO - HbR)
To download the results from the analysis in a .png format:
- Click the Download Data button in the icon menu at the top right of the figure.
Analysis: EEG
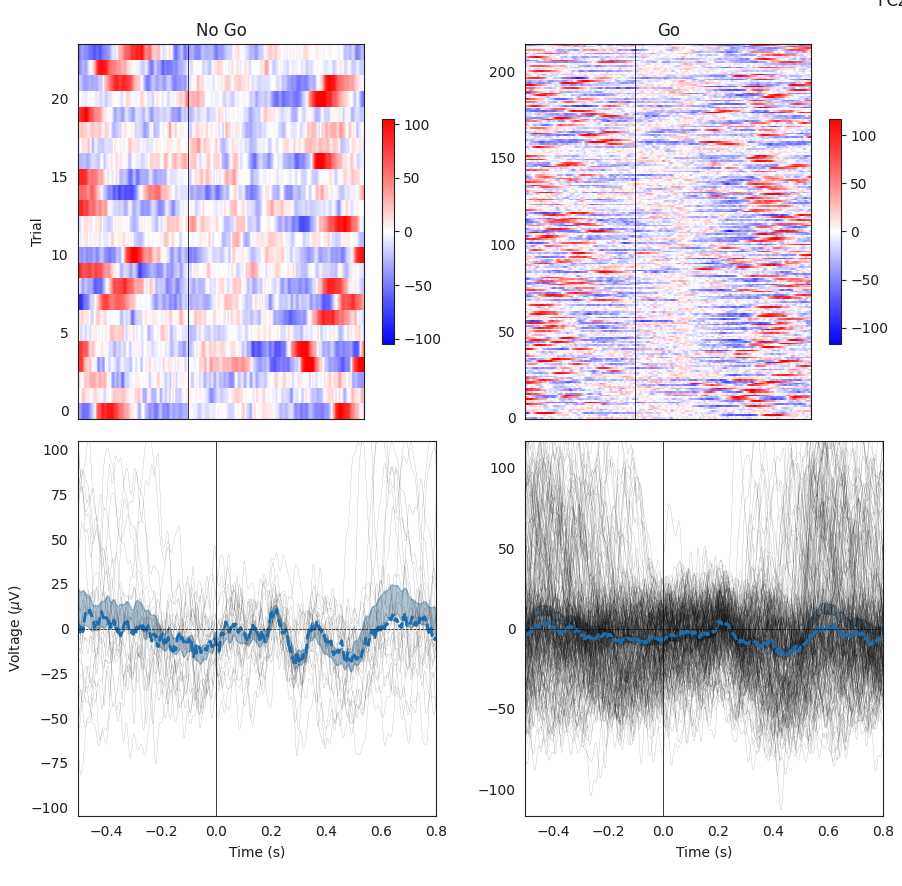
This example Task-specific EEG analysis also comes from the Go/No-Go Task. Any Task with an event-related design will have a similar output. In this case, the two conditions from this Task are separated by column, with time across the x-axis for each trial. In the first row, the trial number is on the y-axis and the voltage value is represented by the heatmap range to the right of each plot. These heatmaps should correspond with the time series data in the second row, where each gray line is a trial and the blue line is the average across all trials.
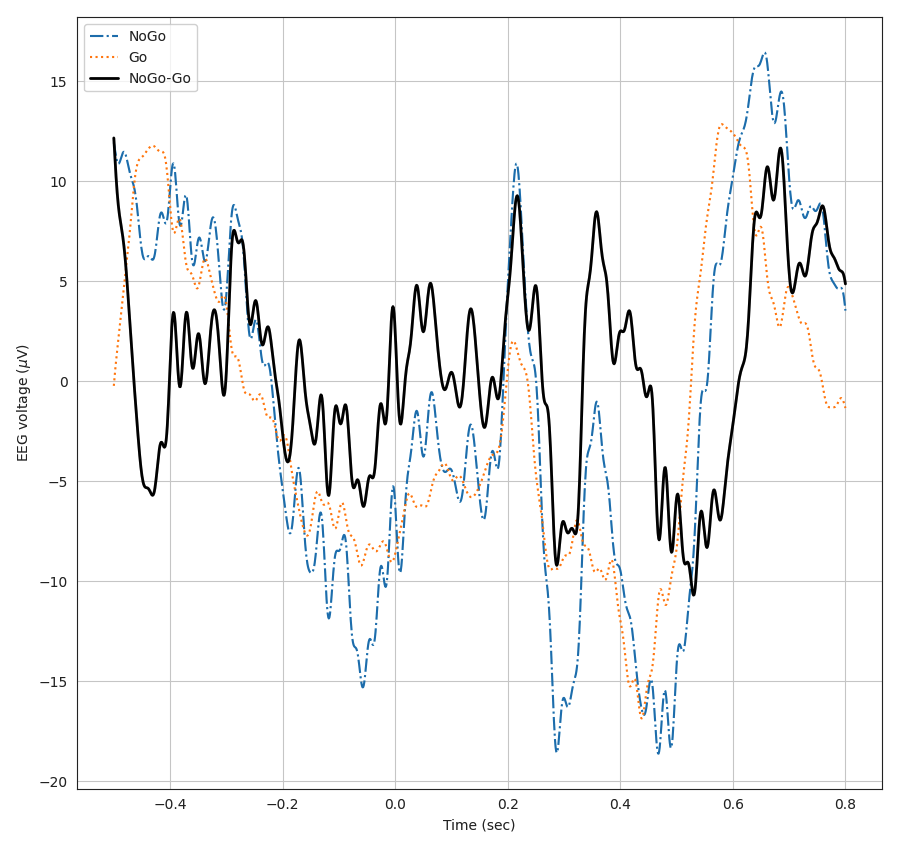 This plot also visualizes the average epoched trial response for EEG data. The two conditions and their difference are specified in the legend at the top-left corner of the plot.
This plot also visualizes the average epoched trial response for EEG data. The two conditions and their difference are specified in the legend at the top-left corner of the plot.