Kernel Flow provides four quality control (QC) reports to help you evaluate the results of your Flow data:
To view or download QC files:
- In the Portal, navigate to the dataset you want to analyze:
- Click Pipelines to open the Pipelines tab.
- Click the QC header to expand that section.
- Click one of the buttons to download the QC file(s).
Descriptions of the QC reports are listed below.
NIRS Basic quality control report
This report contains a Session Summary and several sections, each of which assigns a status to a certain aspect of data quality. The statuses are determined by color and are listed below.
- Red: indicates an issue
- Orange: indicates a warning or potential issues
- Green: indicates no issues
Sections that are assigned orange or red statuses should serve as flags to improve data recording conditions or to retake the dataset.
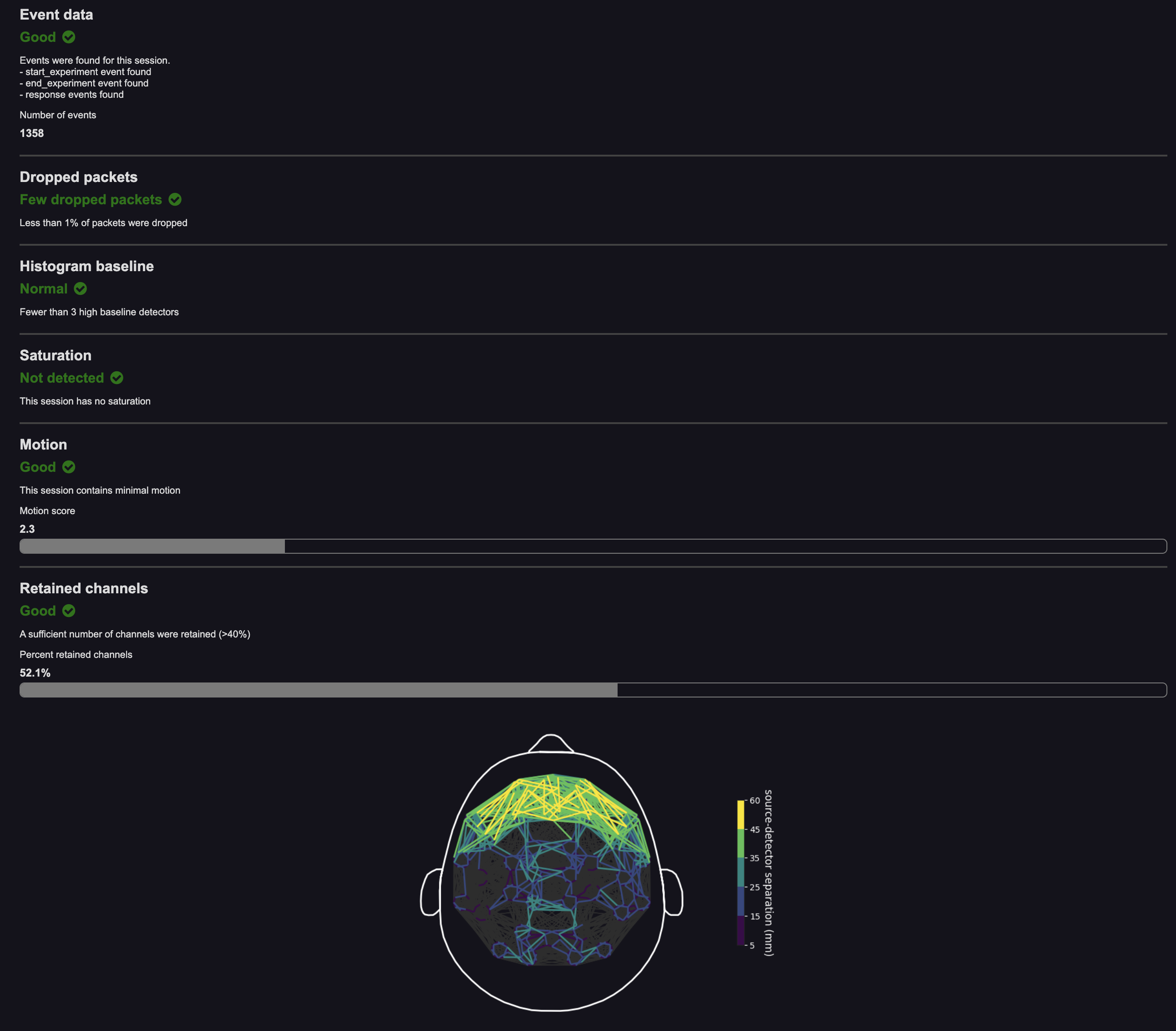
- Event data: This section provides an overview of Task events received for this dataset. It checks for the number of
start_experimentevents,end_experimentevents, and response events (key presses made by the participant). It also lists the number of total events in the session. An orange or red status indicates that a certain type of events was not received. You may ignore this warning if the dataset was not supposed to have Task events or you did not expect certain events, like user responses. - Laser Intensity: This section checks whether the target number of photon counts was achieved on channels with a long source-detector separation (SDS). An orange or red status might indicate hardware issues or failure to tune lasers, which can result in detector saturation.
- This section will only show up in the Basic QC if it is marked with a red or orange status.
- Dropped packets: This section checks for dropped data due to USB transfer or other issues. An orange or red status might indicate software issues.
- This section will only show up in the Basic QC if it is marked with a red or orange status.
- Histogram baseline: This section checks for the presence of bad detectors, which usually have high baseline in their histograms. An orange or red status might indicate hardware issues.
- This section will only show up in the Basic QC if it is marked with a red or orange status.
- Saturation: This section checks for saturation of data in channels with short SDS. Saturation may mean that lasers were not properly tuned prior to recording. An orange or red status might indicate hardware issues or failure to tune lasers.
- This section will only show up in the Basic QC if it is marked with a red or orange status.
- Motion: This section checks for the presence of motion in the data. An orange or red status indicates that a participant moved too much, which introduced artifact into the data. Attempt to reduce participant motion.
- Signal: This section gives a signal strength "score," based on the percent of channels that are analyzable. An orange or red status indicates signal percentage below 50%. Attempt to obtain better signal next time.
NIRS Expert quality control report
This report contains five sections. Click the name in the header of the report to jump directly to that section.
- Grayplot
- Motion
- Stacked Plot
- Total counts Time Series
- Total counts Topoplot
- Physiology (Physio)
- Retained Channels Topoplot
Grayplot
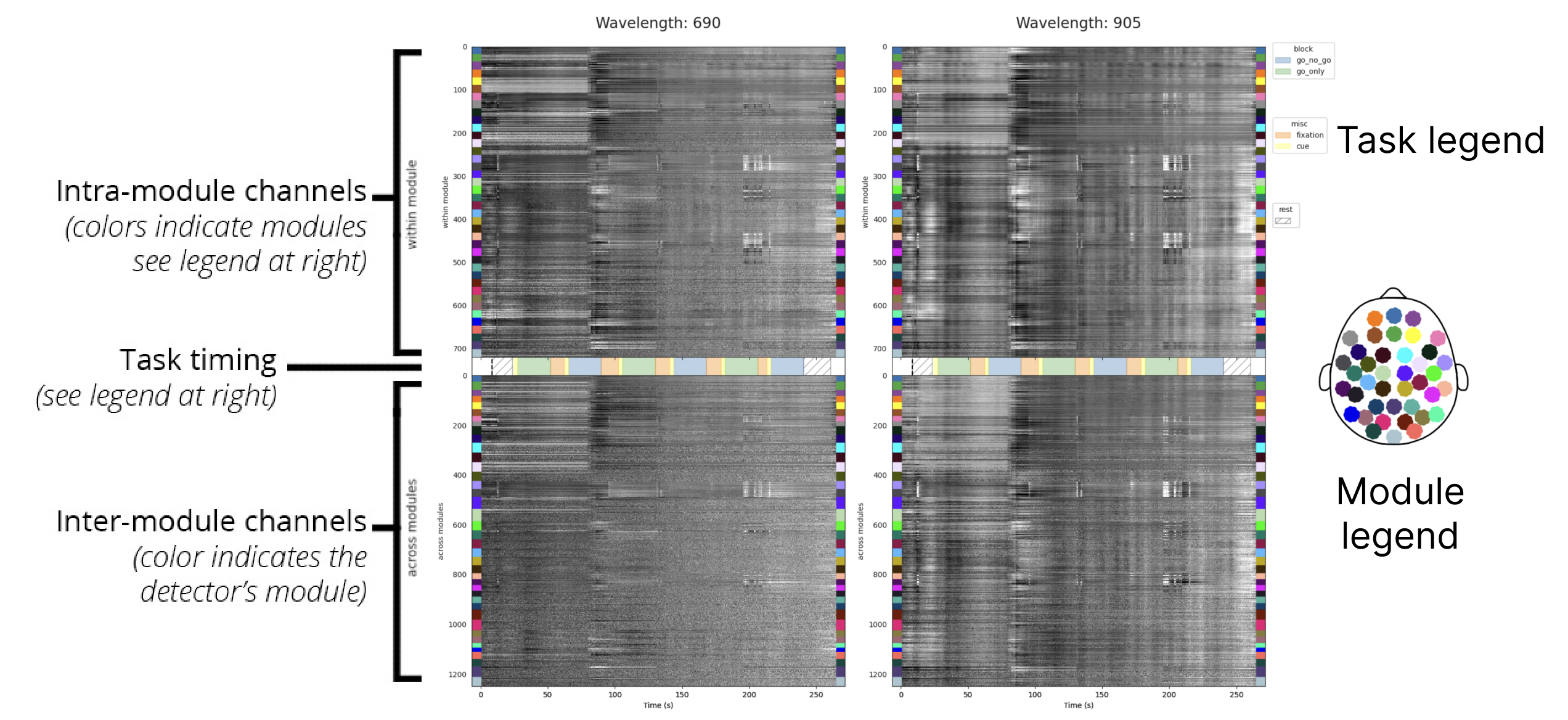
The grayplot (or carpet plot) visualizes global variations in signal intensity in the raw data. To obtain these visualizations, signal fluctuations are normalized (using robust scaling) and depicted with a grayscale color map:
- Signal above the median manifests as lighter regions on the plot.
- Signal below the median corresponds to darker regions.
The top section displays total photon counts recorded from intra-module channels (source and detector in the same module). Data are ordered by module, and color-coded according to the layout overview on the right.
The bottom section displays total photon counts from cross-module channels (source and detector from different modules). This section is color coded based on the module to which the detector belongs.
Interpretation
This visualization highlights global artifacts. Vertical bands or artifacts that span all subplots are indicative of global signal variations, which may correspond to physiological noise, motion, or other non-neural sources (brain activations tend to be somewhat more localized). Artifacts that align with large signal fluctuations in the gyroscopes are likely to be motion-related. Global oscillations that align with the task structure may reflect systemic physiology.
Motion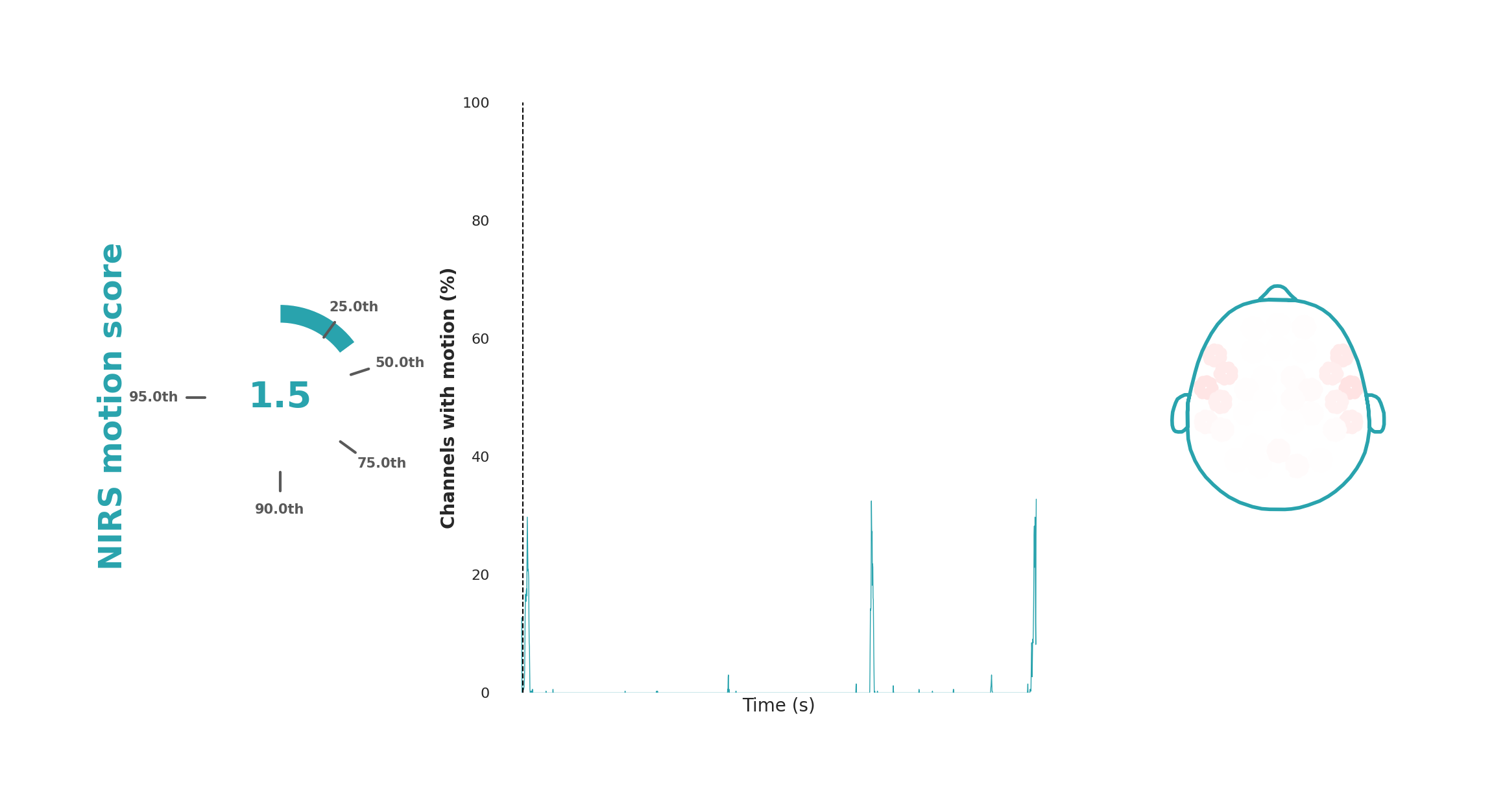
The motion plot visualizes participant head motion during a recording, which introduces noticeable spikes that affect multiple channels simultaneously. For each time point, the number of channels with motion artifacts (spikes) is counted. The middle plot shows the time course of these artifacts, with time on the x-axis and the percentage of affected channels on the y-axis. This plot is overlaid on colored blocks indicating the task structure, if applicable.
To quantify the motion into a score, we average the squared fraction of channels affected by motion during the task blocks, therefore weighting the motion artifacts that affect many channels more heavily. The result is compared to a range observed across 5200 recordings from various studies (Dubois et al., 2024). The percentile in which the current dataset falls is converted to a score from 1 to 10, with 10 indicating the most motion. This score and its percentile are displayed to the left of the time series plot. On the right side, a spatial map shows the percentage of the recording affected by motion per each specific module. For instance, uniform global coloration indicates global head movements, whereas isolated movement in specific regions (e.g., raising eyebrows) would show as localized dark pink or red color.
Interpretation
The motion plot is useful for identifying and assessing the severity of global or region-specific movement artifacts. Kernel's processing pipelines include motion correction, often allowing for the analysis of data with motion spikes. Still, the motion plot provides valuable insights, indicating whether participants need reminders to remain still or if the setup is causing participant discomfort.
Stacked plot
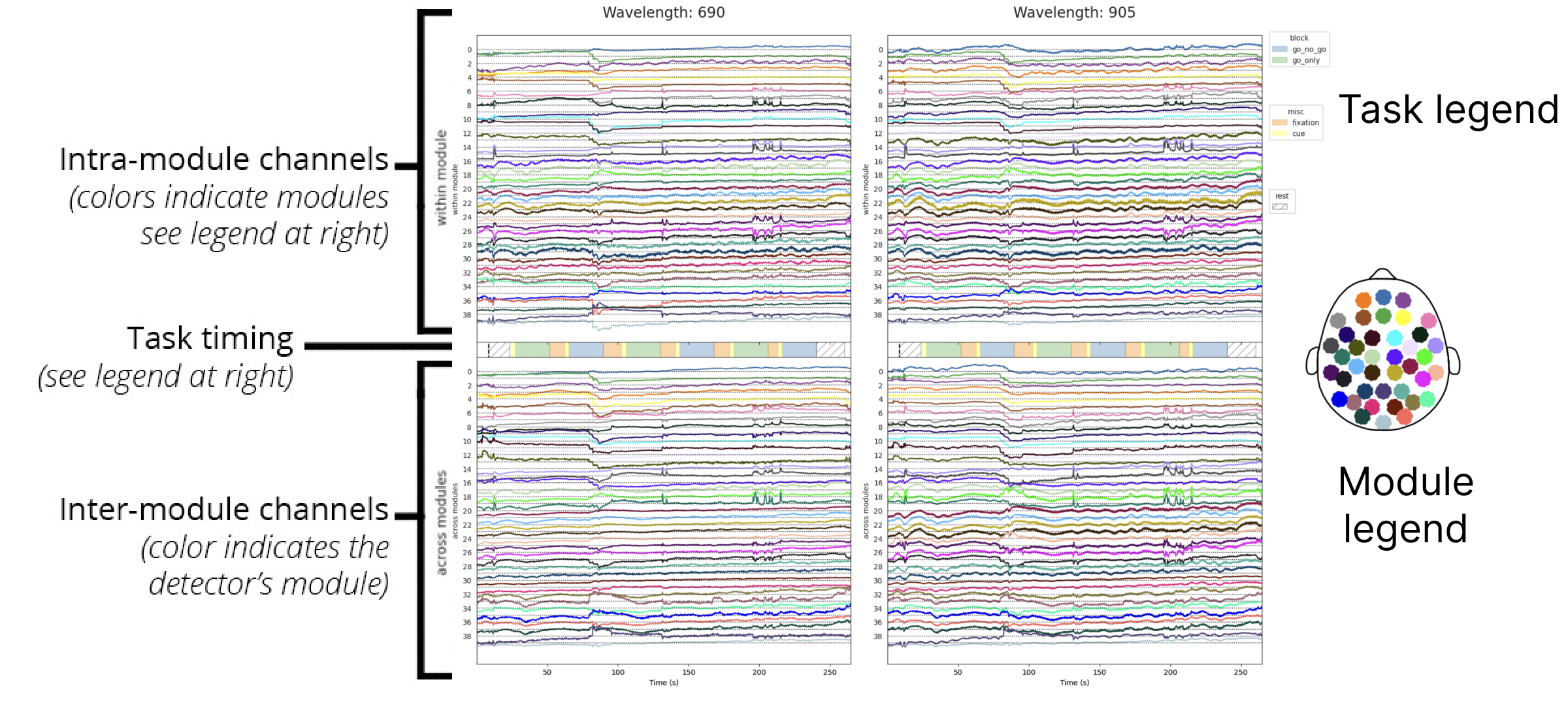
The stacked plot is very similar to the grayplot with an identical layout. The main difference is that the data fluctuations are shown as lines rather than as a heatmap. Also, the data is averaged for all detectors within a module to make the visualization less crowded. For within module channels, signals from the six detectors are averaged together; for across module channels, signals from the six detectors and all incoming sources are averaged together.
Interpretation
Like the grayplot, the stacked plot is useful in identifying global physiological artifacts, movement artifacts, and other non-neural sources. Spike-like events are somewhat easier to identify in this visualization than in the grayplot, making it a good complement.
Total counts time series
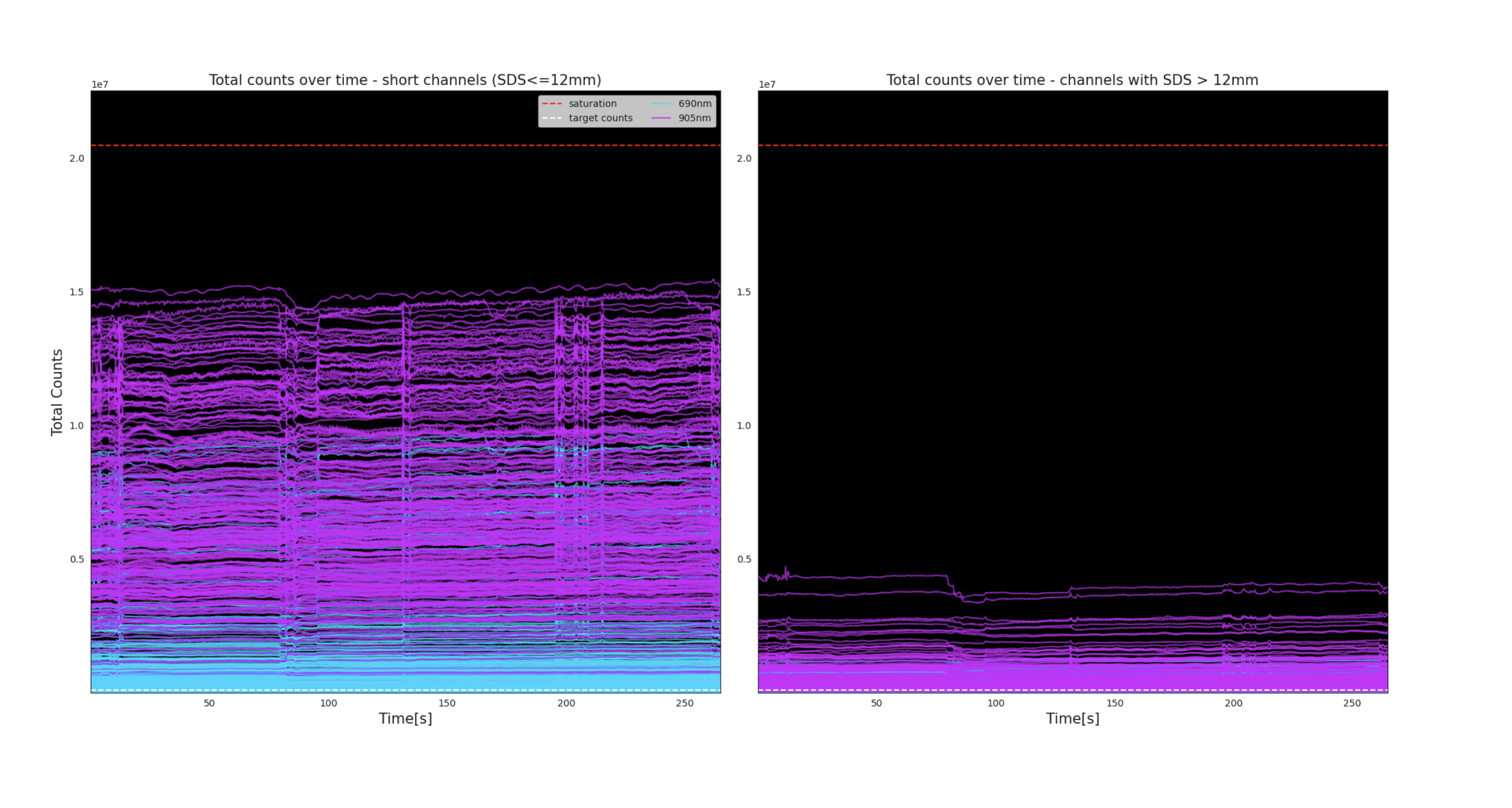
In these plots, total counts for each channel are displayed over the course of the recording. The plot on the left contains within-module channels. The plot on the right contains only across-module channels.
The wavelengths are represented by color: 690nm in cyan and 905nm in magenta. The dashed red horizontal line represents the point of saturation. Saturation may occur if laser power has not been properly adjusted prior to recording (see Tuning the lasers).
Interpretation
This plot provides another method to detect the presence of global artifacts in the data, e.g. large spikes or baseline shifts affecting several channels. This plot is also useful in detecting dead channels or those with poor signal strength, which will show up as lines with consistent low amplitude.
Total Counts topoplot
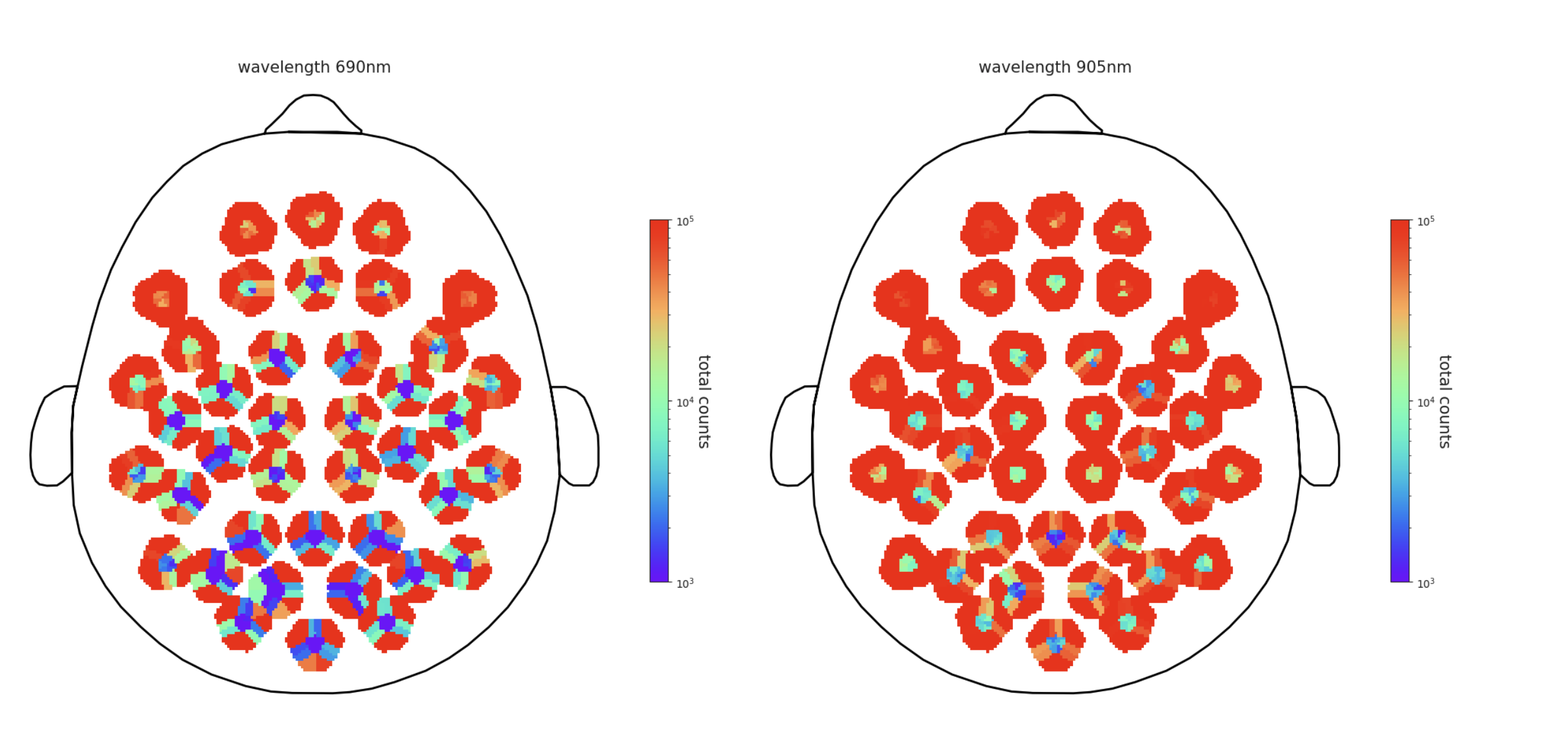
In this figure, mean total counts for all within-module channels are represented spatially as a topoplot. The 690nm wavelength is on the left, and 905nm is on the right. The color map is on a log scale to visualize the range of total counts which may span several orders of magnitude.
Total counts topoplot Interpretation
Total counts are a simple proxy for signal strength. This is similar to the live display in the Kernel Flow Desktop Application, however in this static image the data are averaged across the entire recording.
Physiology (Physio)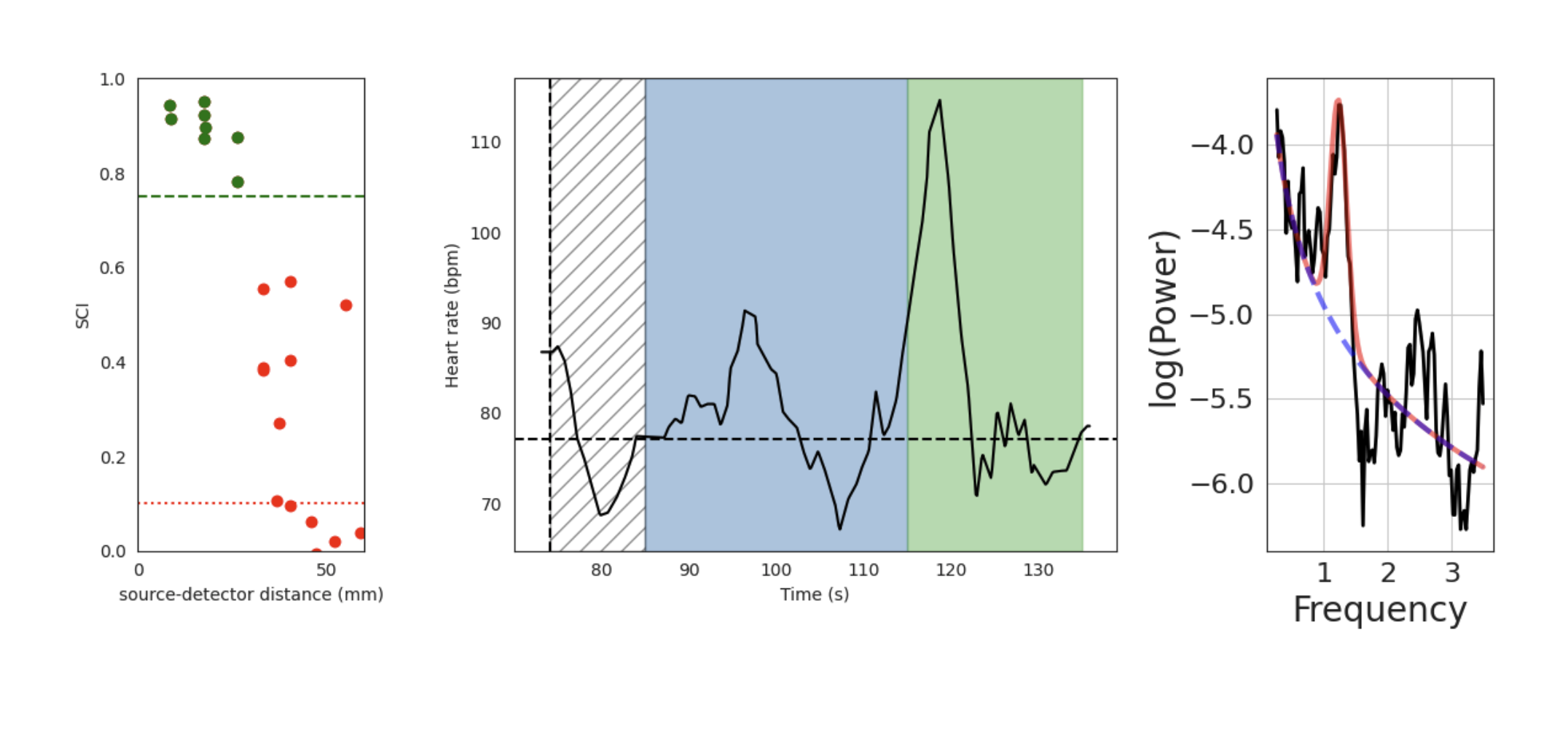
These figures display the physiology metrics pulled from NIRS data. The Flow device typically has several sources which fire at double the rate of all others, which allows us to sample fast enough to capture even the fastest heart rate. On the leftmost graph, each dot represents a channel formed with one of these sources. The x-axis shows the source-detector distance for each channel. The y-axis shows the Scalp Coupling Index (SCI), which measures the correlation between the intensities recorded for the two wavelengths used in our system, after filtering in the heart rate band (0.5 - 2.5 Hz). If it is high—above the green dashed line at 0.75—it is likely that there is a strong heart rate signal. On the middle graph, data from all green channels (the ones with the highest SCI, at least 5) are averaged to create a time by heart rate (in beats per minute) line chart, overlaid on the color-blocked task event sequence (if applicable). On the rightmost graph, the same data is converted to the frequency domain to create a power spectrum. The overlaid red line is fit to the peak of the heart rate based on the black line. The dashed blue line represents the fit to the "aperiodic" component of the power spectrum.
Interpretation
The physiology plots are useful for analyzing data on heart rate during a data recording. If the SCI of the selected channels is low, or the time course of the heart rate is very noisy (large jumps), it may indicate that there was poor contact between the fast firing source and the forehead. Physiological metrics derived from this data may be of poor quality.
Retained Channels topoplot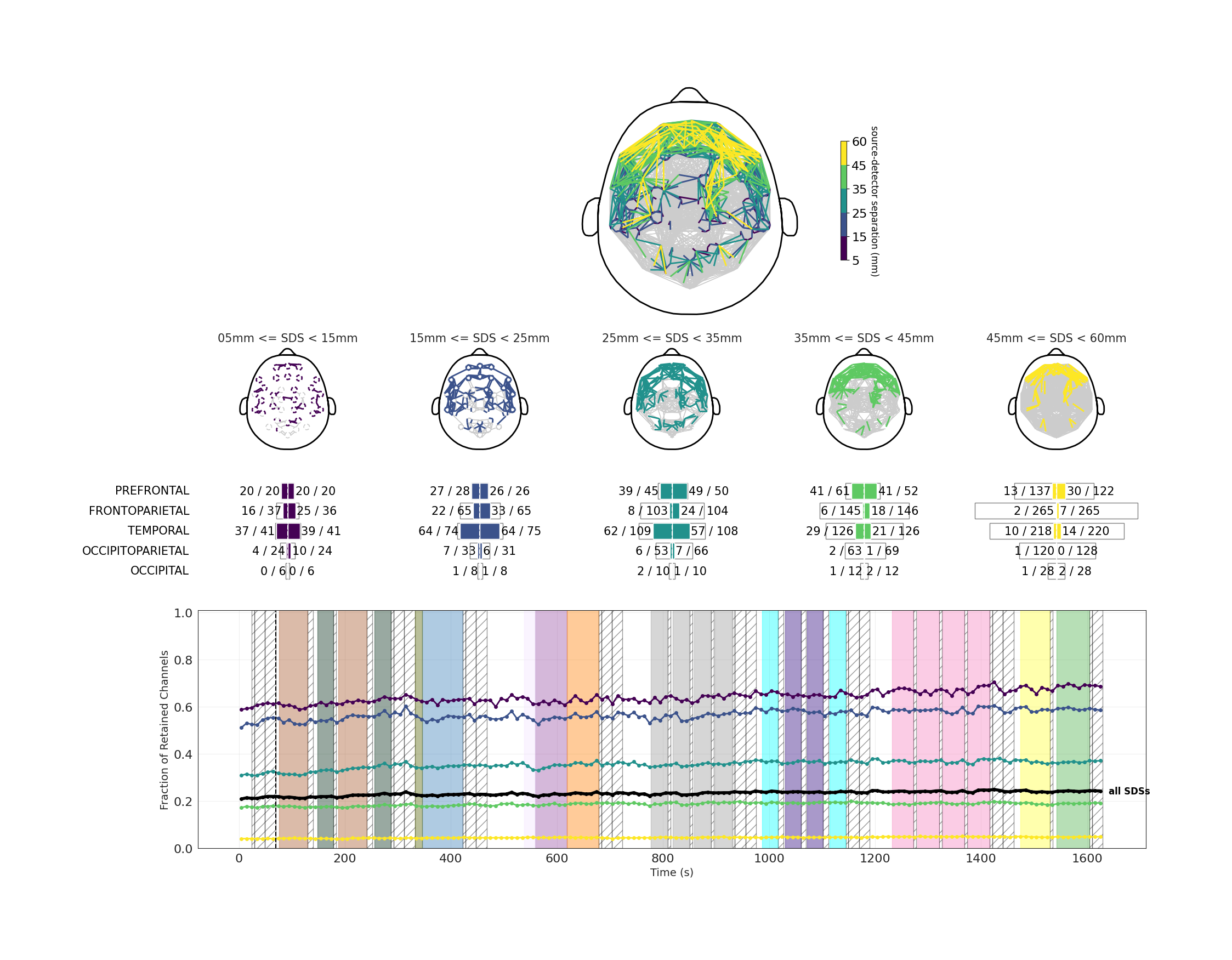
This visualization shows the channels that will be retained for analysis purposes in five different ranges of source-detector separations (SDS). SDS refers to the distance between the light source and the detector for a given channel. The decision for whether a channel is retained takes into account total counts and peak counts, as well as the shape and height of the histograms at the detector.
In the figures, the presence of a color other than gray indicates a channel whose data will be retained for analysis. Lines are drawn between each channel's source and detector. In the top figure, all SDS ranges are overlapped on a topoplot. In the five topoplots just under that, the SDS ranges have been separated by range and color.
In the third row of the visualization, the figures are further quantified by expressing the exact number of channels retained for analysis in each SDS, separated by headset plate. DevKit QC reports will not include this quantification, as the DevKit's module locations are flexible and not divided into plates.
The bottom plot shows how retained channels within each SDS change over time. Each point on the line represents the average percentage of channels retained across the next 10 seconds, with samples containing motion artifacts excluded. Line colors correspond to the same SDS colors used in the topoplots above. In most cases, the number of retained channels increases gradually over time as the device warms up and additional channels cross the inclusion threshold.
Interpretation
Retained channels are a proxy for how much analyzable data was obtained from this participant in this dataset, based on the strength of the signal and the presence of artifacts. You should expect to see higher numbers (and more filled-in topoplots) for the lower ranges of SDS since there is less distance for the light to travel and therefore less risk of the light being blocked in its path. The most important SDS for analysis of NIRS data are 15-25 mm and 25-35 mm. Higher SDS may not have many retained channels, especially in people with thick and/or dark hair.
The bottom plot can also be used to assess the stability of the recording. It shows whether sudden movements or headset adjustments during the session altered the pattern of retained channels. Sessions with large or abrupt changes in the number of retained channels should be interpreted with caution.
EEG quality control report
The EEG QC report is split into four sections. See below for descriptions.
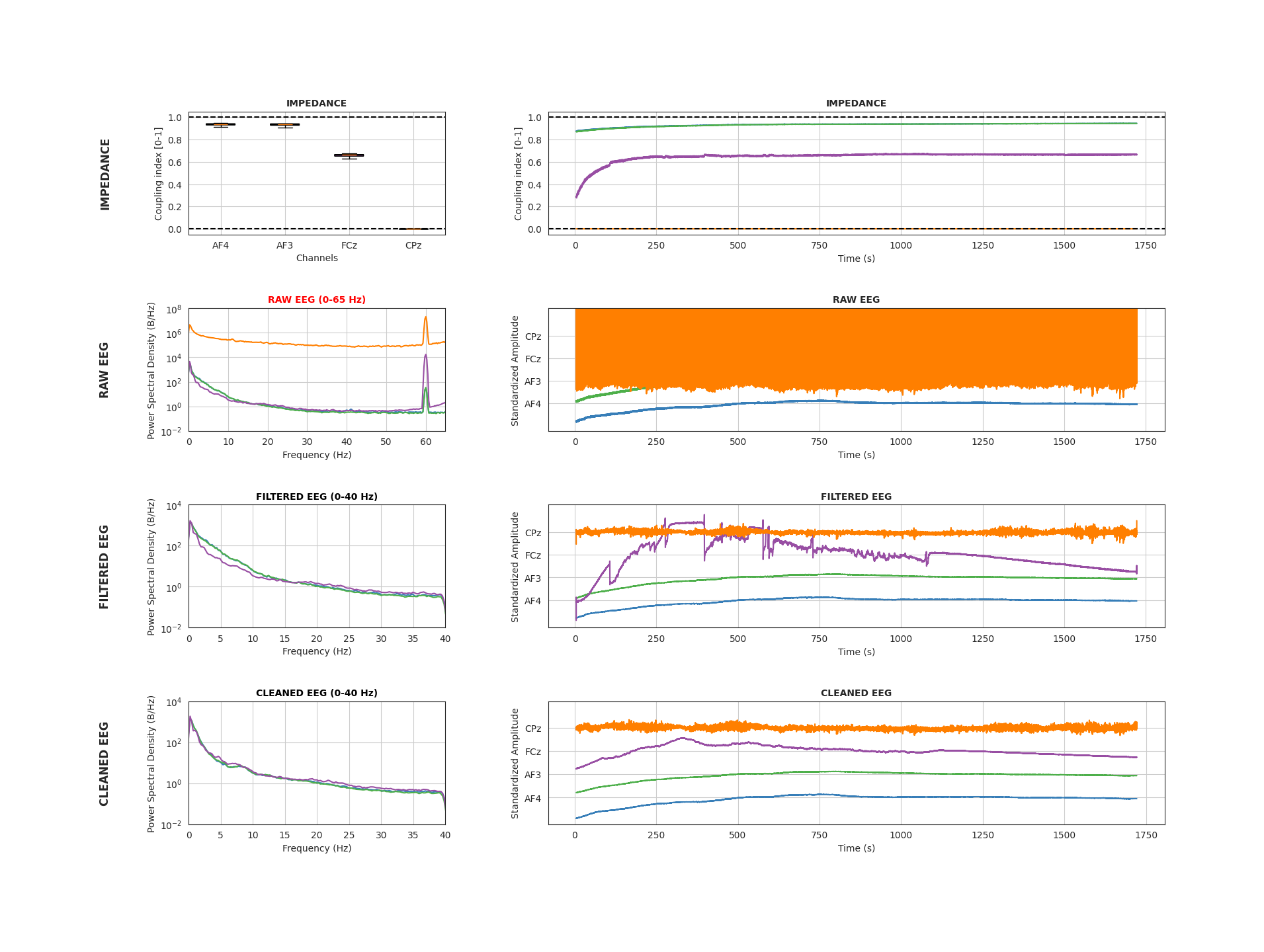
Impedance (Top Row)
A (proprietary) coupling index is computed for each electrode, which varies between 0 and 1, where 1 is the best signal. This index is a proxy for how well "coupled" an electrode is to the scalp. The plot to the left shows the distribution of coupling values over time for each channel, while the plot on the right shows the time course of coupling for each electrode (aligned with the time course of voltages).
Interpretation
Electrodes with better signal will yield less noisy signals, so higher signal strength (>=0.6) is desirable.
Raw EEG (2nd Row)
The plot on the left displays the power spectral density (PSD), showing how EEG signal power is distributed across frequencies from 0 to 65 Hz. This range includes the standard EEG frequency bands as well as the line noise frequency commonly observed in the US (60 Hz). The plot on the right visualizes the unfiltered EEG voltage time course for each channel across the entire recording. Each channel’s time course is stacked along the y-axis.
Interpretation
This plot is useful for identifying noisy channels and assessing the overall quality of the EEG signal, including whether any data are missing. The power spectrum of EEG data typically follows a 1/f distribution, reflecting the aperiodic component of the signal. Peaks are expected in specific frequency bands, most notably around 10 Hz, corresponding to the alpha rhythm, which is especially prominent in occipital electrodes. Spikes at other frequencies—particularly in the higher-frequency range—may indicate noise sources. A distinct peak at the line noise frequency (60 Hz in the US, 50 Hz in Europe) is common and may appear more pronounced in channels with poor electrode contact.
Filtered EEG (3rd Row)
The plots are identical in nature to those in the 2nd row. However, the raw EEG signal has been filtered to the 0.1 - 40 Hz range. Note the change in x-axis limits on the power spectrum.
Interpretation
This plot is useful for assessing overall signal quality after filtering out high-frequency noise that may contaminate the signal.
Cleaned EEG (Bottom Row)
The plots are identical in structure to those shown in the third row. In this case, the filtered EEG signal has undergone additional cleaning using wavelet filtering—a technique designed to remove or attenuate artifacts such as motion, blinks, and other non-neural noise sources.
Interpretation
This plot is useful for assessing overall signal quality following additional data cleaning. It typically reflects the version of the EEG data that will be used for downstream analyses.
Sync Accessory Box quality control report
The Sync Accessory Box QC is only relevant if you purchased a Sync Accessory Box and it was properly plugged into the data acquisition computer during the recording of this dataset.
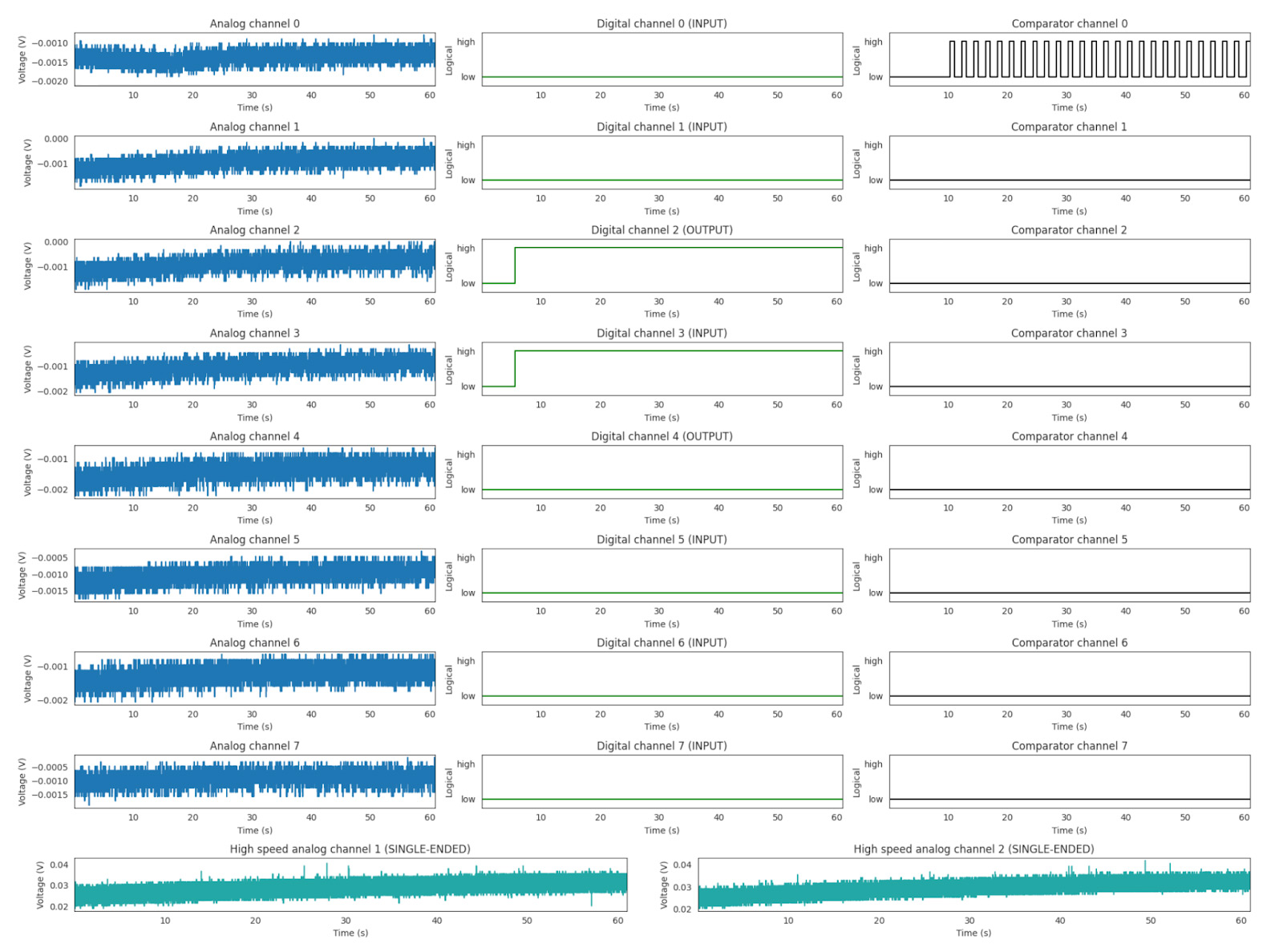
This single multipanel figure displays the outputs from each of the Sync Accessory Box streams as a time series over the course of the experiment. In the analog channels, the data are visualized with a continuous y-axis (voltage). In the digital and comparator channels, a logical “high/low” is used to visualize the data.